Research
We are interested in plant morphogenesis, the process through which plants form new organs. This complex process requires chemical and mechanical information to be integrated across several organisational scales but is remarkably robust despite variability at the cellular scale.
Plant morphogenesis depends critically on growth anisotropy at the cellular scale, which in turn is dominated by mechanical properties of the cell wall surrounding each cell. To gain insights into the highly complex process of morphogenesis, we are employing a multi-disciplinary approach drawing on our expertise in cell and developmental biology, biophysics, and computational mechanics.
Quantitative 4D Confocal Microscopy
Department of Plant Sciences, Charlotte Kirchhelle
We have developed simple imaging chambers in which plant organ development can be followed over several days using confocal microscopy. We use the open-source application MorphoGraphX to extract cell volumes from resulting data sets and calculate cell growth in different directions (Figure 1).
Dynamic Atomic Force Microscopy
Department of Physics, Sonia Contera and Jacob Seifert
We are using dynamic Atomic Force Microscopy (AFM) to map elastic and viscous cell wall properties at the nano scale in living plant tissues (Figure 2) and use the resulting data to develop mechanical models of cell wall behaviour. We also use this system to study fundamental questions about non-equilibrium thermodynamics in multicellular organisms.
Finite Element Models of plant growth
Department of Engineering Science, Antoine Jérusalem, Daniel Garcia-Gonzalez, and Charlotte Kirchhelle
We are using a continuum mechanical framework to develop models of plant growth on the cell, tissue, and organ scale. We use OxFEMM (Oxford Finite Element and Meshless Methods), a bespoke code for modelling of biological tissues undergoing large deformations, to discretise constitutive models (Figure 3).
Current Projects
Leverhulme Early Career Fellowship awarded to Dr. Charlotte Kirchhelle
Summary
How do organisms develop the diverse anatomical shapes observed in nature? Answering this question is pivotal for our basic understanding of multicellular organisms and the rational improvement of domesticated species. However, the underlying process of morphogenesis is highly complex: it involves integrating different kinds of information (genetic, biochemical, biomechanical, and geometric) across multiple scales in space and time. Since this complexity poses a significant challenge to traditional experimental approaches, I will adopt an interdisciplinary approach combining classic molecular biology with state-of-the-art quantitative imaging and computational modelling to study plant morphogenesis, focussing on the recently identified role of cell geometric edges.
BBSRC Responsive Mode Research Grant BB/P01979X/1 awarded to Dr. Ian Moore, Prof. Sonia Contera, Prof. Antoine Jérusalem, and Dr. Charlotte Kirchhelle
Click here for more details.
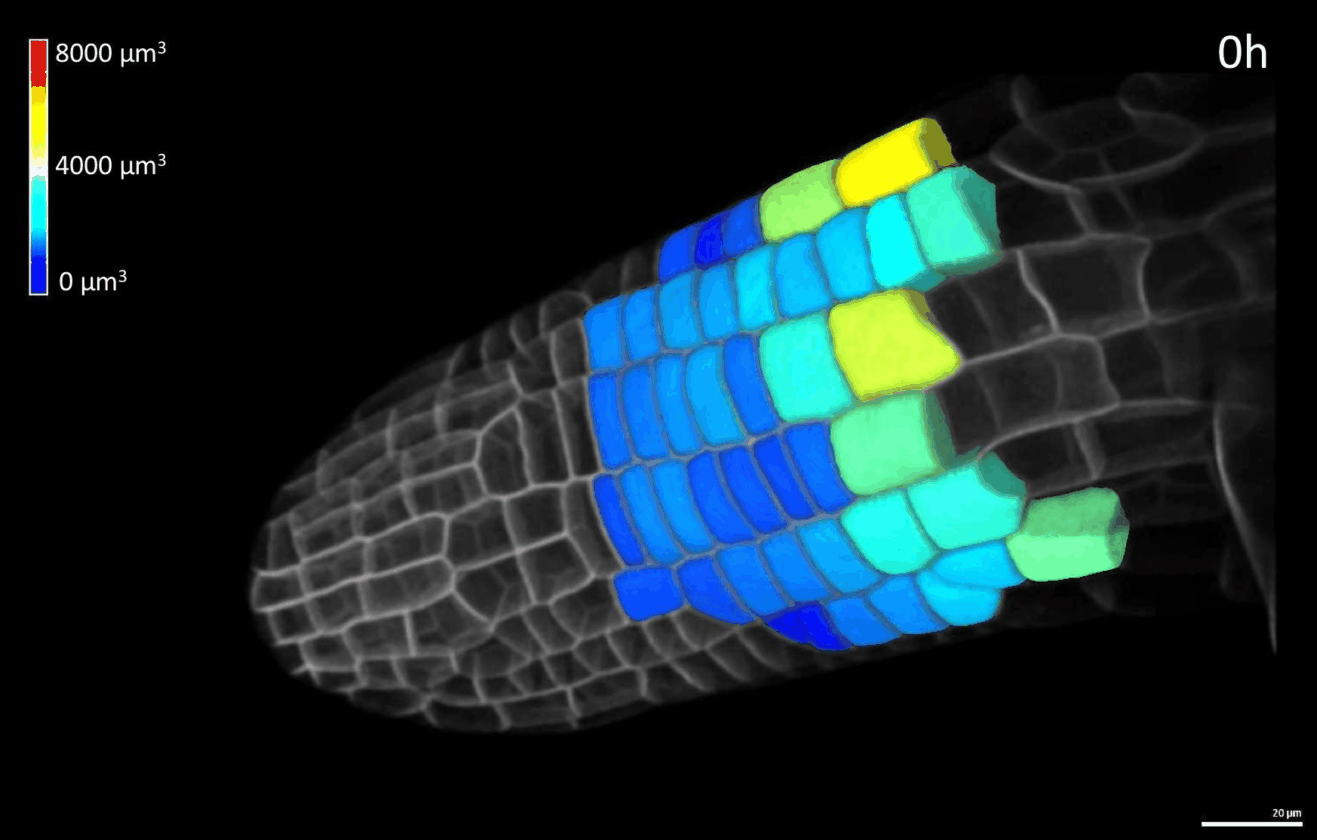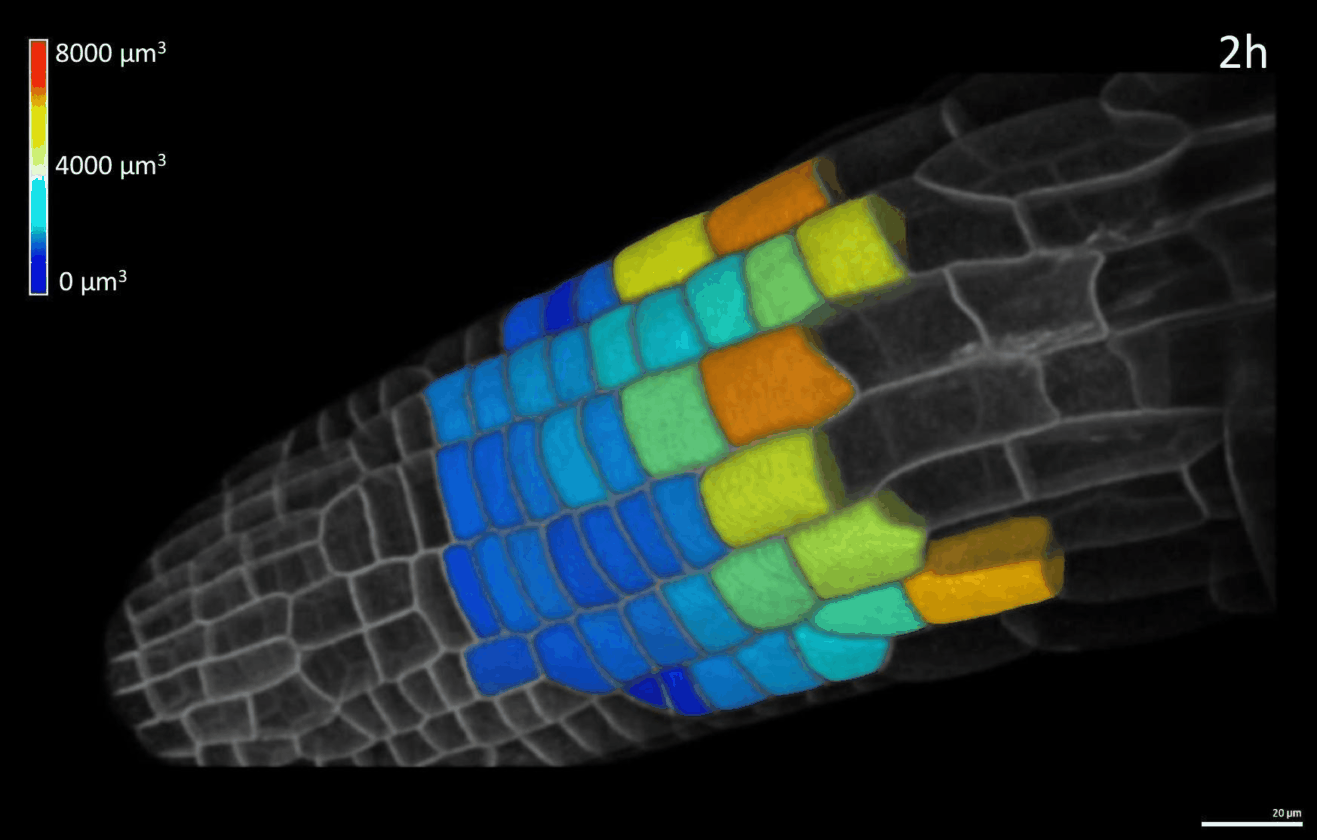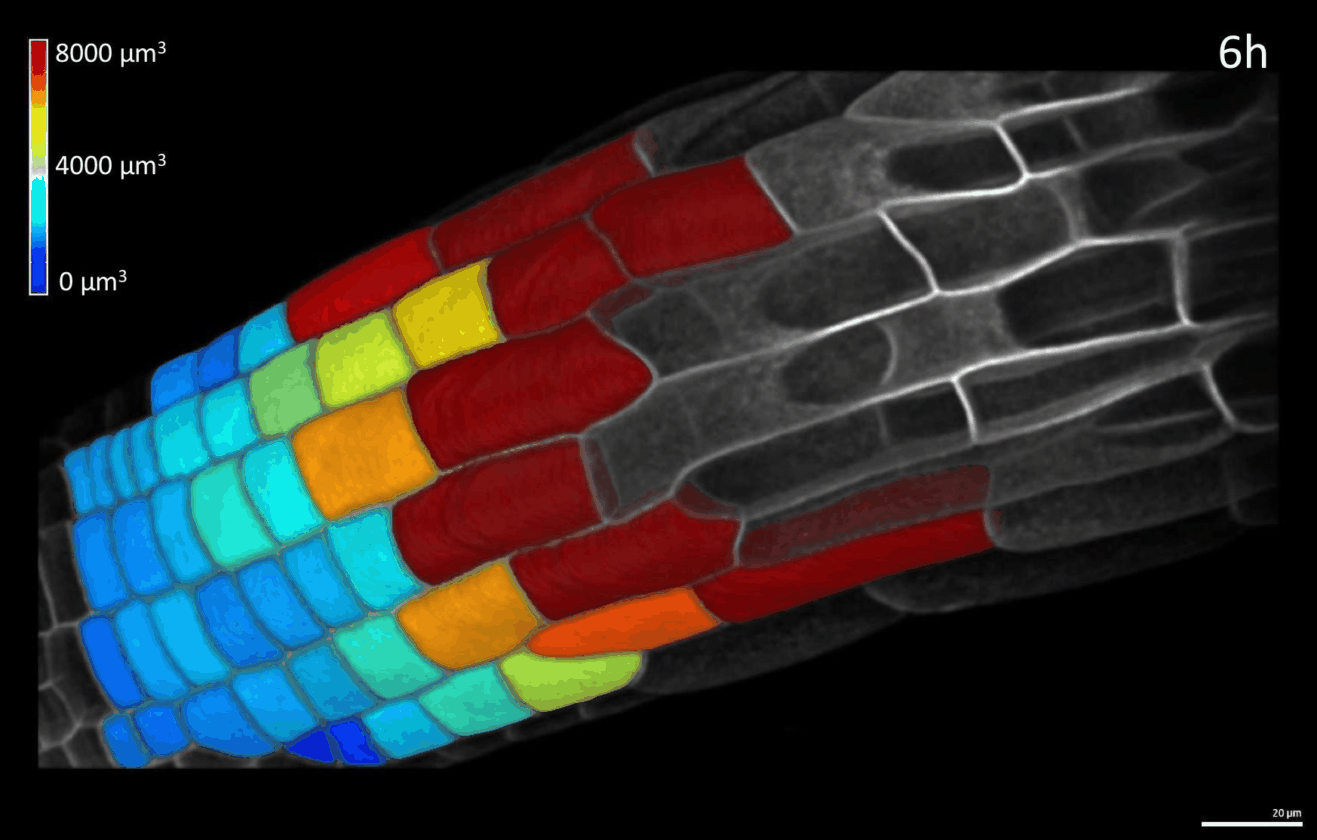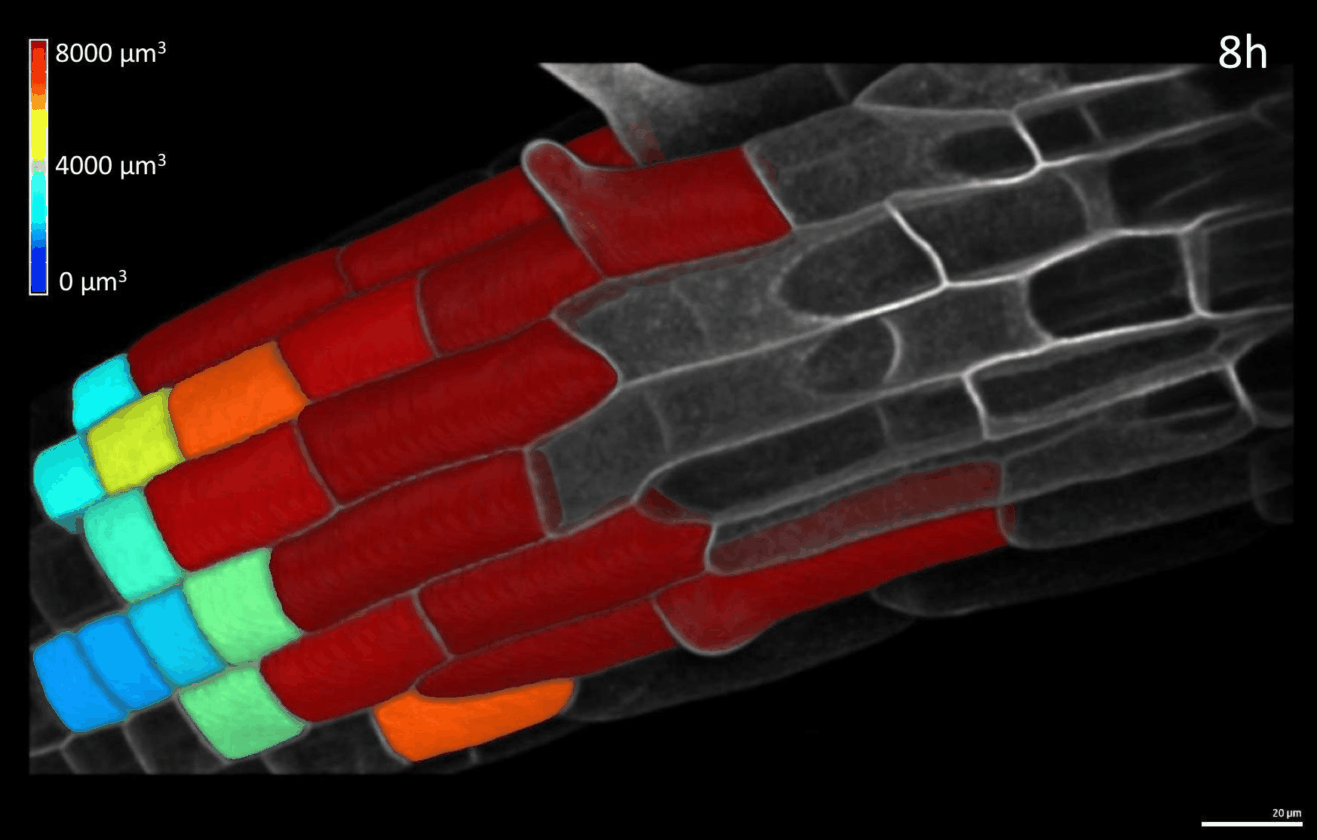
Figure 1: 4D confocal series of a growing root segmented in MorphoGraphX
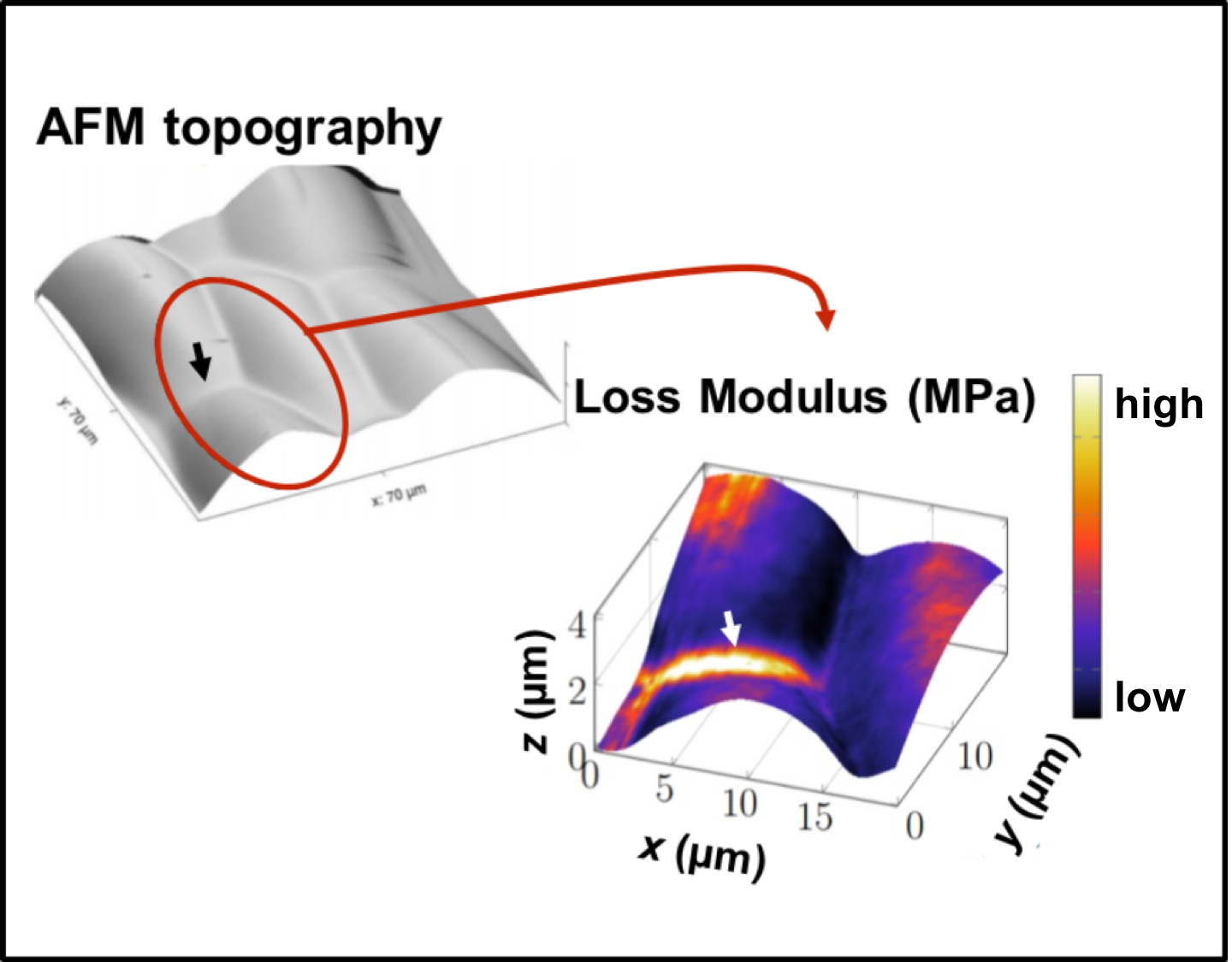
Figure 2: Dynamic AFM of growing plant cells

Figure 3: OxFEMM simulation of an idealised growing plant cell
Interested in joining us?
There are several Doctoral Training Programmes through which students can enter the University. These are fully-funded, four year graduate training programmes which involve a training period of taught courses and two rotation projects before deciding on a DPhil project. Below, we have listed some examples for rotation projects and DPhil projects following on from them. If you are interested in discussing these or other potential projects, please get in touch!
Lead contact: Prof. Antoine Jérusalem
Background. Plants are sessile organisms and thus cannot move, but have developed sophisticated mechanisms to sense their environment and adapt their growing body to it. This developmental flexibility allows them to improve their performance when faced with different environmental stresses. For example, a seedling germinating covered in soil faces very different challenges from one on the soil surface: The seedling buried underground will have to reach the surface to be able to photosynthesise, and has to push through the soil towards the surface in order to achieve this. By contrast, a seedling germinating on the soil surface will be able to photosynthesise, but will be exposed to other environmental factors like wind or heavy rain. The different demands of these environments are reflected in the staggeringly different morphology of plants grown in the absence or presence of light, mimicking these different environments. In the absence of light, plant hypocotyls (the organs connecting root and shoot) rapidly and dramatically elongate to a length up to 10-fold that of hypocotyls developed in the light, which in turn are much wider. These differences in morphology are caused purely by differences in cell growth patterns, which in turn are determined by the mechanical properties of the cell walls surrounding each cell.
While experimental biology has uncovered individual components involved in plant growth and development, studying the mechanics of growing plants and their environment across the multiple spatio-temporal scales involved in plant development remains a key challenge. How does a plant respond to different mechanical environments? How do cell wall mechanical properties change, and what effect do these changes have on growth? Computational modelling has the capacity to integrate quantitative mechanical information of growing plants and their surroundings across multiple scales, and is thus able to solve this fundamental challenge in plant biology.
Aims:
- Develop a computational model of hypocotyl development. Finite Element (FE) models of hypocotyls will be developed using the group code OxFEMM (Oxford Finite Element and Meshless Methods). Models will be informed by 3D confocal images of hypocotyls.
- Explore the effect of mechanically different growth environments on hypocotyl development in silico. Different growth media will be mechanically characterised in indendation experiments, resulting data will be used to inform in silico hypocotyl growth through different substrates.
Using a simple experimental system, this project will develop a model capable of interrogating the influence of external factors on emergent properties of developing organs. This project provides a starting point to explore how changes at the cellular and subcellular level can change organ-level properties. During a subsequent DPhil project, this work will be extended to develop meso-scale models incorporating emergent properties from small-scale events. This work can then be extended into other organs and species to inform rational strategies to improve mechanical performance of plants.
Lead contact: Prof. Sonia Contera
Background: Growth is as fundamental to life as replication: without it a replicating organism would just become smaller and smaller. Growth of organisms is governed by the dynamic mechanical properties of biological structures, which arise from their molecular composition and structure, interactions with the environments and chemical activity. Mechanical properties are time dependent, because they emerge from the capacity of biological materials to store (elasticity) and dissipate (viscosity) mechanical energy, in other words, viscoelasticity determines biological shapes and their mechanical behaviour across temporal and spatial scales, from the molecular to the cellular to the organ level.
In recent years we have developed techniques based on the atomic force microscope (AFM) that have allowed us to map for the first time the viscoelastic properties of live cells in living organisms with nanometre resolution. We are using this ability not only to link growth and shape to the biological activity of cells, but also to understand the use of energy in off- equilibrium biological process in the context of eukaryotic morphogenesis, and tissue engineering among other applications.
In this project we are particularly interested in the combination of genetic, biochemical, mechanical, and geometric factors across multiple scales in space and time in plant morphogenesis. In plants, morphogenesis is dominated by the cell wall, which determines each cell’s polyhedral form and their relative position in the tissue. Cellular growth in cylindrical organs like roots is largely unidirectional, however organs with more complex geometries (e.g. leaves and flowers) require multi-directional growth. Cellular growth rate and direction depend on the precisely targeted deposition and modification of cell wall macromolecules (principally cellulose and pectins) at different faces of the cell. But this is not enough. Recent research (led by collaborator Charlotte Kirchhelle) has shown that cells require an additional mechanism: identification of their geometric edges, a process dominated by a small GTPase protein (RAB-A5c).
Aims: This project proposes to investigate the question: Why do plants possess t two independent mechanisms to control cellular growth direction, and how do these mechanisms relate to each other?
We propose to use the AFM to determine the relation between shape, local and global energy dissipated and stored, and local (nanoscale) and global (cellular and organ level) time response of cells during leaf morphogenesis.
Future DPhil project: This short project would lead to a DPhil as part of a multidisciplinary collaboration across departments that aims at understanding biological morphogenesis. Possibilities include (but are not restricted to) (1) mapping cell wall mechanical properties at the subcellular scale and investigate functional links to cell- and organ-level behaviour, using state-of-the-art AFM (with Prof. Contera, Physics), (2) a largely computational project developing quantitative models of plant organ development (with Prof. Jérusalem, Engineering Science), (3) investigating the molecular machinery involved in growth control through a genetic screen for suppressors and enhancers of the RAB-A5c phenotype (with Dr Kirchhelle, Plant Sciences).
Lead contact: Dr. Charlotte Kirchhelle
Background: How do multicellular organisms generate the astonishing morphological diversity found in nature? The underlying process of morphogenesis is highly complex and involves the integration of genetic, biochemical, mechanical, and geometric factors across multiple scales in space and time. In plants, morphogenesis is dominated by the rigid cell wall, which also determines each plant cell’s polyhedral form and fixes cells in their relative position in the tissue. Morphogenesis consequently requires groups of cells to coordinate their growth rates and directions. While cellular growth in cylindrical organs like roots is largely unidirectional, organs with more complex geometries, like leaves and flowers, require multi-directional cellular growth. Cellular growth rate and direction depend on the precisely targeted deposition and modification of cell wall macromolecules (principally cellulose and pectins) at different faces of the cell. We recently reported that such cells can also identify their geometric edges (where two faces meet) as a distinct spatial domain, and showed that the plant-specific small GTPase RAB-A5c specifies an apparently selective exocytic trafficking pathway to cell edges. Disruption of this pathway dramatically shifted cell growth direction from typically unidirectional (anisotropic) to near isotropic growth, causing severe morphological defects at the cell-, tissue-, and organ-scale. Recent in vivo and in silico data indicate that RAB-A5c-mediated control of cellular growth direction is independent of the previously described mechanism regulating growth direction through oriented deposition of cellulose at different cell faces (facial anisotropy) in unidirectionally growing lateral roots.
Why do plants possess these two independent mechanisms to control cellular growth direction, and how do these mechanisms relate to each other? We hypothesise these mechanisms act either on different time-scales or in different directions to fine-tune growth and facilitate robust morphogenesis. Due to its single growth vector, it is challenging to investigate the spatio-temporal relationship between RAB-A5c activity and facial anisotropy in lateral roots. We therefore propose to use leaf morphogenesis, which depends on multidirectional growth, as an experimental system to test the questions laid out above.
Aims:
- Establish a 4D imaging and quantitative image analysis pipeline to quantify cellular growth during leaf development.
- Dissect the spatio-temporal relationship of facial anisotropy and RAB-A5c activity during leaf development.
Future DPhil project: This short project provides a starting point for a DPhil project in different areas, depending on the student’s interest. Possibilities include (1) a largely computational project developing quantitative models of plant organ development investigating the role of subcellular features like cell edges on the organ-scale (with Prof. Antoine Jerusalem, Department of Engineering Science), (2) mapping cell wall mechanical properties at the subcellular scale and investigate functional links to cell- and organ-level behaviour, using state-of-the-art atomic force microscopy (with Prof. Sonia Contera, Department of Physics), (3) investigating the molecular machinery involved in RAB-A5c-mediated growth control through a genetic screen for suppressors and enhancers of the RAB-A5c phenotype, (4) investigating the evolutionary importance of cell edges during plant morphogenesis through investigating RAB-A5 function in basal land plants lineages (liverworts and mosses).


